Metop-B IASI - Total Column Carbon Monoxide - Level 2
Contents
Metop-B IASI - Total Column Carbon Monoxide - Level 2¶
The Infrared Atmospheric Sounding Interferometer (IASI) is an instrument onboard the Metop-B/C satellites. It was also previously aboard Metop-A. It provides information on the vertical structure of temperature and humidity as well as main atmospheric species.
This notebook provides you an introduction to data from Metop-B IASI. This dataset is used as a proxy for data from IASI-NG, a passive infrared sounder which has the capability to measure the temperature and water vapour profiles of the Earth’s atmosphere. IASI Level 2 data can be downloaded from the IASI portal.
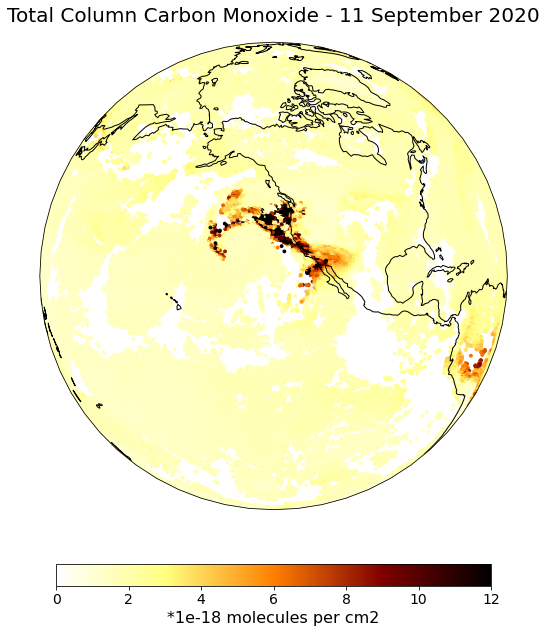
The event featured is the August Complex fire in California, USA in 2020. This was the largest wildfire in CA history, spreading over 1,000,000 acres (over 4,000 sq km).
Basic Facts
Spatial resolution: 4 x 12-km coverage close to the centre of a 48 x 48 km2 cell (average sampling distance: 24 km)
Spatial coverage: Near global
Revisit time: less than one day
Data availability: since 2007
How to access the data
IASI Level 2 are disseminated in the netCDF format and can be downloaded via the IASI portal.
Load required libraries
import os
import xarray as xr
from datetime import datetime
import numpy as np
from netCDF4 import Dataset
import pandas as pd
# Python libraries for visualization
import matplotlib.pyplot as plt
import matplotlib.colors
import matplotlib.cm as cm
from matplotlib.axes import Axes
import cartopy
import cartopy.crs as ccrs
from cartopy.mpl.gridliner import LONGITUDE_FORMATTER, LATITUDE_FORMATTER
from cartopy.mpl.geoaxes import GeoAxes
GeoAxes._pcolormesh_patched = Axes.pcolormesh
from skimage import exposure
import warnings
warnings.filterwarnings('ignore')
warnings.simplefilter(action = "ignore", category = RuntimeWarning)
Load helper functions
%run ../functions.ipynb
Load and browse IASI Total Column Carbon Monoxide data¶
You can use the Python library xarray to access and manipulate datasets in netCDF format.
Xarray’s function xr.open_dataset enables you to open a netCDF file. Once loaded, you can inspect the data structure.
You see, that the data is a 3-dimensional vector with more than 540,000 individual entries for the time dimension. latitude, longitude and other parameters are stored as individual data variables.
iasi_file = xr.open_dataset('../data/iasi/co/2020/09/11/IASI_METOPB_L2_CO_20200911_ULB-LATMOS_V6.5.0.nc')
iasi_file
<xarray.Dataset>
Dimensions: (time: 542455, nlayers: 19, npressures: 20)
Coordinates:
* time (time) float64 4.322e+08 ... 4.323e+08
Dimensions without coordinates: nlayers, npressures
Data variables: (12/21)
time_string (time) |S16 ...
time_in_day (time) float64 ...
latitude (time) float32 ...
longitude (time) float32 ...
solar_zenith_angle (time) float32 ...
satellite_zenith_angle (time) float32 ...
... ...
CO_total_column (time) float32 ...
CO_total_column_error (time) float32 ...
CO_degrees_of_freedom (time) float32 ...
air_partial_column_profile (time, nlayers) float32 ...
atmosphere_pressure_grid (time, npressures) float32 ...
averaging_kernel_matrix (time, nlayers, nlayers) float32 ...
Attributes: (12/31)
title: IASI/METOPB ULB-LATMOS carbon monoxide (CO) ...
institution: ULB-LATMOS for algorithm development ; EUMET...
product_version: 6.5.0
history: 2020-09-12 06:32:03 (date of data extraction...
summary: This dataset contains Level 2 carbon monoxid...
source: EUMETSAT IASI Level 2 carbon monoxide (CO) d...
... ...
sensor: IASI
spatial_resolution: 12km at nadir
creator_type: institution
creator_name: ULB-LATMOS
contact_email: contact form at http://iasi.aeris-data.fr/co...
data_policy: see https://iasi.aeris-data.fr/data-use-policy/- time: 542455
- nlayers: 19
- npressures: 20
- time(time)float644.322e+08 4.322e+08 ... 4.323e+08
- long_name :
- UTC observation time in seconds since 2007-01-01 00:00:00 UTC
- units :
- second
- standard_name :
- time
array([4.321728e+08, 4.321728e+08, 4.321728e+08, ..., 4.322592e+08, 4.322592e+08, 4.322592e+08])
- time_string(time)|S16...
- long_name :
- UTC observation time as YYYYMMDDThhmmssZ
[542455 values with dtype=|S16]
- time_in_day(time)float64...
- long_name :
- UTC observation time in seconds in the day
- units :
- second
[542455 values with dtype=float64]
- latitude(time)float32...
- standard_name :
- latitude
- long_name :
- latitude of ground pixel center
- units :
- degrees_north
- valid_range :
- [-90. 90.]
[542455 values with dtype=float32]
- longitude(time)float32...
- standard_name :
- longitude
- long_name :
- longitude of ground pixel center
- units :
- degrees_east
- valid_range :
- [-180. 180.]
[542455 values with dtype=float32]
- solar_zenith_angle(time)float32...
- long_name :
- solar zenith angle at the Earth s surface for the pixel center
- units :
- degrees
- standard_name :
- solar_zenith_angle
[542455 values with dtype=float32]
- satellite_zenith_angle(time)float32...
- long_name :
- Metop zenith angle at the Earth s surface for the pixel center
- units :
- degrees
- standard_name :
- platform_zenith_angle
[542455 values with dtype=float32]
- orbit_number(time)int64...
- long_name :
- Metop orbit number
[542455 values with dtype=int64]
- scanline_number(time)int32...
- long_name :
- scanline number in the Metop orbit
[542455 values with dtype=int32]
- pixel_number(time)int32...
- long_name :
- pixel number in the current scanline
- valid_range :
- [ 1 120]
[542455 values with dtype=int32]
- ifov_number(time)int32...
- long_name :
- field of view number in the 2 x 2 observation matrix
- valid_range :
- [1 4]
[542455 values with dtype=int32]
- retrieval_quality_flag(time)int32...
- long_name :
- retrieval quality flag summarizing processing flags
- comment :
- = 2 for the most reliable pixels; = 1 for the valuable pixels, to use with caution; = 0 for the remaining pixels that we recommend not to use
[542455 values with dtype=int32]
- surface_altitude(time)float32...
- units :
- m
- long_name :
- altitude of the surface
- standard_name :
- surface_altitude
[542455 values with dtype=float32]
- CO_apriori_partial_column_profile(time, nlayers)float32...
- units :
- mol m-2
- long_name :
- carbon monoxide a priori partial column vertical profile in mole/m2 in the layers defined by the levels given in the variable atmosphere_pressure_grid
- standard_name :
- mole_content_of_ozone_in_atmosphere_layer
- comment :
- When the first levels are not available (because of the orography), CO_apriori_partial_column_profile = -999. for these levels.
- multiplication_factor_to_convert_to_molecules_per_cm2 :
- 6.02214086e+19
[10306645 values with dtype=float32]
- CO_partial_column_profile(time, nlayers)float32...
- units :
- mol m-2
- long_name :
- carbon monoxide partial column vertical profile in mole/m2 retrieved in the layers defined by the levels given in the variable atmosphere_pressure_grid
- standard_name :
- mole_content_of_carbon_monoxide_in_atmosphere_layer
- comment :
- When the first levels are not available (because of the orography), CO_partial_column_profile = -999. for these levels.
- ancillary_variables :
- CO_partial_column_error
- multiplication_factor_to_convert_to_molecules_per_cm2 :
- 6.02214086e+19
[10306645 values with dtype=float32]
- CO_partial_column_error(time, nlayers)float32...
- long_name :
- vertical profile of total retrieval error associated to carbon monoxide partial column vertical profile in the layers defined by the levels given in the variable atmosphere_pressure_grid
- comment :
- This error is relative to the carbon monoxide total column; it is without unit.
- standard_name :
- atmosphere_mole_content_of_carbon_monoxide standard_error
[10306645 values with dtype=float32]
- CO_total_column(time)float32...
- units :
- mol m-2
- long_name :
- retrieved carbon monoxide total column in mole/m2
- standard_name :
- atmosphere_mole_content_of_carbon_monoxide
- ancillary_variables :
- CO_total_column_error
- multiplication_factor_to_convert_to_molecules_per_cm2 :
- 6.02214086e+19
[542455 values with dtype=float32]
- CO_total_column_error(time)float32...
- long_name :
- total retrieval error associated to carbon monoxide total column
[542455 values with dtype=float32]
- CO_degrees_of_freedom(time)float32...
- long_name :
- degrees of freedom of the signal in the retrieved carbon monoxide partial column profile
- comment :
- This is the estimation of the number of independant information pieces concerning carbon monoxide vertical distribution in the signal.
[542455 values with dtype=float32]
- air_partial_column_profile(time, nlayers)float32...
- units :
- mol m-2
- long_name :
- air partial column vertical profile in mole/m2 in the layers defined by the levels given in the variable atmosphere_pressure_grid
- comment :
- When the first levels are not available (because of the orography), air_partial_column_profile = -999. for these levels.
- standard_name :
- mole_content_of_air_in_atmosphere_layer
- multiplication_factor_to_convert_to_molecules_per_cm2 :
- 6.02214086e+19
[10306645 values with dtype=float32]
- atmosphere_pressure_grid(time, npressures)float32...
- units :
- Pascal
- long_name :
- pressures in Pa corresponding to levels used to define inversion layers: 18 layers of about 1 km height between Earth s surface and 18 km with an additional layer from 18 km to the top of the atmosphere (60 km)
- comment :
- When the first levels are not available (because of the orography), atmosphere_pressure_grid = -999. for these levels. When the pressure profile could not be estimated, atmosphere_pressure_grid = -999. for all levels.
- standard_name :
- air_pressure
[10849100 values with dtype=float32]
- averaging_kernel_matrix(time, nlayers, nlayers)float32...
- long_name :
- carbon monoxide partial column averaging kernels matrix ((mol/m2)/(mol/m2)) in the layers defined by the levels given in the variable atmosphere_pressure_grid
- comment :
- When the first levels are not available (because of the orography), averaging_kernel_matrix = -999. for these levels.
[195826255 values with dtype=float32]
- title :
- IASI/METOPB ULB-LATMOS carbon monoxide (CO) L2 products (profiles of partial columns and total columns)
- institution :
- ULB-LATMOS for algorithm development ; EUMETSAT for data production ; LATMOS for data reconstruction and formatting ; AERIS for data access
- product_version :
- 6.5.0
- history :
- 2020-09-12 06:32:03 (date of data extraction) - Product generated with FORLI v20151001 at EUMETSAT
- summary :
- This dataset contains Level 2 carbon monoxide profile and total column products from IASI observations. These products were generated at EUMETSAT under the auspices of AC SAF with the ULB-LATMOS FORLI algorithm version 20151001 using the EUMETSAT v8.0 IASI Level 1C and v6.5 IASI Level 2 temperature, humidity and cloud data. They are distributed in files in BUFR format via EumetCast. Data reconstruction and formatting in netcdf files are processed by LATMOS. Quicklooks and data access are provided by AERIS
- source :
- EUMETSAT IASI Level 2 carbon monoxide (CO) data version 6.5
- references :
- Reference to the CO retrieval: FORLI radiative transfer and retrieval code for IASI, J. Quant. Spectrosc. Ra., 113, 1391-1408, https://doi.org/10.1016/j.jqsrt.2012.02.036, 2012.
- id :
- IASI_METOPB_L2_CO_20200911_ULB-LATMOS_V6.5.0.nc
- tracking_id :
- e7d9ad0e-f4b0-11ea-acff-73d7f7916c85
- geospatial_lat_min :
- -90.0
- geospatial_lat_max :
- +90.0
- geospatial_latitude_units :
- degrees_north
- geospatial_lon_min :
- -180.0
- geospatial_lon_max :
- +180.0
- geospatial_longitude_units :
- degrees_east
- geospatial_vertical_min :
- 0
- geospatial_vertical_max :
- 60
- geospatial_vertical_units :
- km
- time_coverage_start :
- 20191201T000000Z
- time_coverage_end :
- 20191201T235959Z
- conventions :
- CF-1.6
- standard_name_vocabulary :
- NetCDF Climate and Forecast (CF) Medata Convention version 73, 23 June 2020
- keywords :
- satellite,observation,atmosphere,carbon monoxide,CO,level 2,column,altitude,profile,pollution,IASI,Metop-B
- keywords_vocabulary :
- GCMD Science Keywords
- platform :
- Metop-B
- sensor :
- IASI
- spatial_resolution :
- 12km at nadir
- creator_type :
- institution
- creator_name :
- ULB-LATMOS
- contact_email :
- contact form at http://iasi.aeris-data.fr/contact/
- data_policy :
- see https://iasi.aeris-data.fr/data-use-policy/
Retrieve the variable ‘CO total column’ as xarray.DataArray¶
You can specify one variable of interest by putting the name of the variable into square brackets [] and get more detailed information about the variable. E.g. CO_total_column , which holds data values for total column carbon monoxide.
co = iasi_file['CO_total_column']
co
<xarray.DataArray 'CO_total_column' (time: 542455)>
[542455 values with dtype=float32]
Coordinates:
* time (time) float64 4.322e+08 4.322e+08 ... 4.323e+08 4.323e+08
Attributes:
units: mol m-2
long_name: retrieved carbon ...
standard_name: atmosphere_mole_c...
ancillary_variables: CO_total_column_e...
multiplication_factor_to_convert_to_molecules_per_cm2: 6.02214086e+19- time: 542455
- ...
[542455 values with dtype=float32]
- time(time)float644.322e+08 4.322e+08 ... 4.323e+08
- long_name :
- UTC observation time in seconds since 2007-01-01 00:00:00 UTC
- units :
- second
- standard_name :
- time
array([4.321728e+08, 4.321728e+08, 4.321728e+08, ..., 4.322592e+08, 4.322592e+08, 4.322592e+08])
- units :
- mol m-2
- long_name :
- retrieved carbon monoxide total column in mole/m2
- standard_name :
- atmosphere_mole_content_of_carbon_monoxide
- ancillary_variables :
- CO_total_column_error
- multiplication_factor_to_convert_to_molecules_per_cm2 :
- 6.02214086e+19
Load data into a xarray.DataArray with the function generate_xr_from_1d_vec¶
With the help of the function generate_xr_from_1d_vec, you can generate a xarray.DataArray object, with latitude and longitude values as coordinates and the total column carbon monoxide information as data values. This data structure will be helpful for plotting and masking the data.
Further, you can retrieve the information for the variables long_name, unit and variable name from the attributes of the data object co.
iasi_co_da = generate_xr_from_1d_vec(file=iasi_file,
lat_path='latitude',
lon_path='longitude',
variable=co.data,
parameter_name=co.name,
longname=co.long_name,
no_of_dims=1,
unit=co.units)
iasi_co_da
<xarray.DataArray 'CO_total_column' (ground_pixel: 542455)>
array([0.03949106, 0.03213775, 0.04679925, ..., 0.03180185, 0.03522488,
0.03115139], dtype=float32)
Coordinates:
latitude (ground_pixel) float32 8.665 8.915 8.798 ... 84.91 84.73 85.22
longitude (ground_pixel) float32 -47.95 -48.01 -48.49 ... -66.97 -54.55
Dimensions without coordinates: ground_pixel
Attributes:
long_name: retrieved carbon monoxide total column in mole/m2
units: mol m-2- ground_pixel: 542455
- 0.03949 0.03214 0.0468 0.04191 ... 0.03435 0.0318 0.03522 0.03115
array([0.03949106, 0.03213775, 0.04679925, ..., 0.03180185, 0.03522488, 0.03115139], dtype=float32) - latitude(ground_pixel)float328.665 8.915 8.798 ... 84.73 85.22
array([ 8.664803, 8.915398, 8.798302, ..., 84.9109 , 84.7296 , 85.2249 ], dtype=float32) - longitude(ground_pixel)float32-47.95 -48.01 ... -66.97 -54.55
array([-47.949097, -48.0121 , -48.4937 , ..., -68.1499 , -66.9676 , -54.545998], dtype=float32)
- long_name :
- retrieved carbon monoxide total column in mole/m2
- units :
- mol m-2
Mask Total Column Carbon Monoxide data by quality flag¶
Load quality flag information¶
The IASI Level 2 data files provide you information on the quality for each data point. This information is useful to generate a quality mask and to mask out data points with a non sufficient quality.
In order to do so, you have to load the quality flag variable retrieval_quality_flag from the data file. The pixels with a quality flag = 2 are the most reliable pixels. Pixels with a quality flag = 1 or 0 shall be masked out.
qf = iasi_file['retrieval_quality_flag']
qf
<xarray.DataArray 'retrieval_quality_flag' (time: 542455)>
[542455 values with dtype=int32]
Coordinates:
* time (time) float64 4.322e+08 4.322e+08 ... 4.323e+08 4.323e+08
Attributes:
long_name: retrieval quality flag summarizing processing flags
comment: = 2 for the most reliable pixels; = 1 for the valuable pixels...- time: 542455
- ...
[542455 values with dtype=int32]
- time(time)float644.322e+08 4.322e+08 ... 4.323e+08
- long_name :
- UTC observation time in seconds since 2007-01-01 00:00:00 UTC
- units :
- second
- standard_name :
- time
array([4.321728e+08, 4.321728e+08, 4.321728e+08, ..., 4.322592e+08, 4.322592e+08, 4.322592e+08])
- long_name :
- retrieval quality flag summarizing processing flags
- comment :
- = 2 for the most reliable pixels; = 1 for the valuable pixels, to use with caution; = 0 for the remaining pixels that we recommend not to use
You can re-use the generate_xr_from_1d_vec function again in order to generate a xarray.DataArray with the quality flag information.
iasi_co_qf_da = generate_xr_from_1d_vec(file=iasi_file,
lat_path='latitude',
lon_path='longitude',
variable=qf,
parameter_name=qf.name,
longname=qf.long_name,
no_of_dims=1,
unit='-')
iasi_co_qf_da
<xarray.DataArray 'retrieval_quality_flag' (ground_pixel: 542455)>
array([1, 1, 1, ..., 2, 2, 2], dtype=int32)
Coordinates:
latitude (ground_pixel) float32 8.665 8.915 8.798 ... 84.91 84.73 85.22
longitude (ground_pixel) float32 -47.95 -48.01 -48.49 ... -66.97 -54.55
Dimensions without coordinates: ground_pixel
Attributes:
long_name: retrieval quality flag summarizing processing flags
units: -- ground_pixel: 542455
- 1 1 1 1 2 1 2 2 2 1 2 2 1 1 1 2 2 ... 2 2 1 2 1 2 2 2 1 2 1 1 1 2 2 2
array([1, 1, 1, ..., 2, 2, 2], dtype=int32)
- latitude(ground_pixel)float328.665 8.915 8.798 ... 84.73 85.22
array([ 8.664803, 8.915398, 8.798302, ..., 84.9109 , 84.7296 , 85.2249 ], dtype=float32) - longitude(ground_pixel)float32-47.95 -48.01 ... -66.97 -54.55
array([-47.949097, -48.0121 , -48.4937 , ..., -68.1499 , -66.9676 , -54.545998], dtype=float32)
- long_name :
- retrieval quality flag summarizing processing flags
- units :
- -
Mask the Total Column Carbon Monoxide data¶
The quality flag information can now be used to mask the xarray.DataArray with the data values. You can make use of the function generate_masked_array, where you can specify which pixels shall remain and which ones shall be eliminated. All data points with a quality flag = 2 shall be kept, all others shall be masked out.
You see that the number of data points reduced to just a bit less than 400,000 instead of more than 500,000.
iasi_co_masked = generate_masked_array(xarray=iasi_co_da,
mask=iasi_co_qf_da,
threshold=2,
operator='=')
iasi_co_masked
<xarray.DataArray (ground_pixel: 392634)>
array([0.02875579, 0.0296423 , 0.03025403, ..., 0.03180185, 0.03522488,
0.03115139], dtype=float32)
Coordinates:
latitude (ground_pixel) float32 9.989 10.37 10.54 ... 84.91 84.73 85.22
longitude (ground_pixel) float32 -43.75 -41.56 -41.6 ... -66.97 -54.55
Dimensions without coordinates: ground_pixel
Attributes:
long_name: retrieved carbon monoxide total column in mole/m2
units: mol m-2- ground_pixel: 392634
- 0.02876 0.02964 0.03025 0.02999 ... 0.02866 0.0318 0.03522 0.03115
array([0.02875579, 0.0296423 , 0.03025403, ..., 0.03180185, 0.03522488, 0.03115139], dtype=float32) - latitude(ground_pixel)float329.989 10.37 10.54 ... 84.73 85.22
array([ 9.989197, 10.371399, 10.535301, ..., 84.9109 , 84.7296 , 85.2249 ], dtype=float32) - longitude(ground_pixel)float32-43.75 -41.56 ... -66.97 -54.55
array([-43.745903, -41.5605 , -41.599 , ..., -68.1499 , -66.9676 , -54.545998], dtype=float32)
- long_name :
- retrieved carbon monoxide total column in mole/m2
- units :
- mol m-2
Convert units of Total Column Carbon Monoxide data¶
The last step before visualizing the total column carbon monoxide information is to convert the data from mol/m2 to molecules/cm2. The loaded data variable co has an attribute called multiplication_factor_to_convert_to_molecules_per_cm2, which is used to convert the data values.
iasi_co_masked_converted = iasi_co_masked*co.multiplication_factor_to_convert_to_molecules_per_cm2
iasi_co_masked_converted
<xarray.DataArray (ground_pixel: 392634)>
array([1.7317139e+18, 1.7851012e+18, 1.8219404e+18, ..., 1.9151522e+18,
2.1212919e+18, 1.8759807e+18], dtype=float32)
Coordinates:
latitude (ground_pixel) float32 9.989 10.37 10.54 ... 84.91 84.73 85.22
longitude (ground_pixel) float32 -43.75 -41.56 -41.6 ... -66.97 -54.55
Dimensions without coordinates: ground_pixel- ground_pixel: 392634
- 1.732e+18 1.785e+18 1.822e+18 ... 1.915e+18 2.121e+18 1.876e+18
array([1.7317139e+18, 1.7851012e+18, 1.8219404e+18, ..., 1.9151522e+18, 2.1212919e+18, 1.8759807e+18], dtype=float32) - latitude(ground_pixel)float329.989 10.37 10.54 ... 84.73 85.22
array([ 9.989197, 10.371399, 10.535301, ..., 84.9109 , 84.7296 , 85.2249 ], dtype=float32) - longitude(ground_pixel)float32-43.75 -41.56 ... -66.97 -54.55
array([-43.745903, -41.5605 , -41.599 , ..., -68.1499 , -66.9676 , -54.545998], dtype=float32)
Visualize IASI Total Column Carbon Monoxide¶
You can visualize the IASI Total Column Carbon Monoxide data with the function visualize_scatter, which uses matplotlib’s scatterplot function. You can set an Orthographic() projection and focus on a region over California.
visualize_scatter(xr_dataarray=iasi_co_masked_converted,
conversion_factor=1e-18,
projection=ccrs.Orthographic(-130,30),
vmin=0,
vmax=12,
point_size=8,
color_scale='afmhot_r',
unit='*1e-18 molecules per cm2',
title='Total Column Carbon Monoxide - 11 September 2020')
References¶
IASI is a joint mission of EUMETSAT and the Centre National d’Etudes Spatiales (CNES, France). The authors acknowledge the AERIS data infrastructure for providing access to the IASI data in this study, ULB-LATMOS for the development of the retrieval algorithms, and Eumetsat/AC SAF for CO data production
Some code in this notebook was adapted from the following source:
copyright: 2022, EUMETSAT
license: MIT
retrieved: 2022-06-28 by Sabrina Szeto
Return to the case study
Monitoring smoke transport with next-generation satellites from Metop-SG: Californian Wildfires Case Study
EPS-SG IASI-NG Carbon Monoxide
