Sentinel-3 OLCI Composites
Contents
Sentinel-3 OLCI Composites¶
Sentinel-3 OLCI Level-1 data products are available as Full (OL_1_EFR) and Reduced Resolution (OL_1_ERR) data files. The following notebook shows you how OL_1_EFR data are structured and how you can generate true- and false-colour composite images.
Find more information on the OLCI data products in the Sentinel-3 OLCI User Guide.
This notebook provides an introduction to the Sentinel-3 OLCI data and enables you to generate true- and false-colour composite images of the fires in southern Italy and in Greece that occured in August 2021.
Basic Facts
Spatial resolution: 300m
Spatial coverage: Global
Revisit time: Less than 2 days
Data availability: since 2016
How to access the data
Sentinel-3 OLCI data can be downloaded via the Copernicus Open Access Hub. You need to register for an account before downloading data.
Sentinel-3 OLCI data products are disseminated as .zip archives, containing data files in NetCDF format.
Load required libraries
import os
import xarray as xr
import numpy as np
import netCDF4 as nc
import pandas as pd
import glob
from IPython.display import HTML
# Python libraries for visualization
%matplotlib inline
import matplotlib.pyplot as plt
import matplotlib.colors
from matplotlib.cm import get_cmap
from matplotlib.axes import Axes
import cartopy.crs as ccrs
from cartopy.mpl.gridliner import LONGITUDE_FORMATTER, LATITUDE_FORMATTER
import cartopy.feature as cfeature
from cartopy.mpl.geoaxes import GeoAxes
GeoAxes._pcolormesh_patched = Axes.pcolormesh
from skimage import exposure
import warnings
warnings.simplefilter(action = "ignore", category = RuntimeWarning)
warnings.simplefilter(action = "ignore", category = UserWarning)
Load helper functions
%run ../functions.ipynb
Load and browse Sentinel-3 OLCI Level-1 data¶
Sentinel-3 Level-1 data are dissiminated as .zip archives when downloaded.
[OPTIONAL] The first step is to unzip file from the zipped archive downloaded. This is optional as we have already unzipped the file for you. This is why the code is commented out.
# import zipfile
# with zipfile.ZipFile('../data/sentinel-3/olci/2021/08/07/S3B_OL_1_EFR____20210807T085200_20210807T085500_20210808T130135_0179_055_278_2340_LN1_O_NT_002.zip', 'r') as zip_ref:
# zip_ref.extractall('../data/sentinel-3/olci/2021/08/07/')
The unzipped folder contains 30 data files in NetCDF format. Data for each channel is stored in a single NetCDF file. Additionally, you get information on qualityFlags, time_coordinates or geo_coordinates.
You can see the names of the 30 data files by looping through the data directory. You see that the channel information follow the same naming and all end with _radiance.nc.
olci_dir = '../data/sentinel-3/olci/2021/08/07/S3B_OL_1_EFR____20210807T085200_20210807T085500_20210808T130135_0179_055_278_2340_LN1_O_NT_002.SEN3/'
for i in glob.glob(olci_dir+'*.nc'):
tmp = i.split('/')
print(tmp[8])
time_coordinates.nc
Oa01_radiance.nc
Oa02_radiance.nc
Oa03_radiance.nc
Oa04_radiance.nc
Oa05_radiance.nc
Oa06_radiance.nc
Oa07_radiance.nc
Oa08_radiance.nc
Oa09_radiance.nc
Oa10_radiance.nc
Oa11_radiance.nc
Oa12_radiance.nc
Oa13_radiance.nc
Oa14_radiance.nc
Oa15_radiance.nc
Oa16_radiance.nc
Oa17_radiance.nc
Oa18_radiance.nc
Oa19_radiance.nc
Oa20_radiance.nc
Oa21_radiance.nc
qualityFlags.nc
instrument_data.nc
removed_pixels.nc
geo_coordinates.nc
tie_geo_coordinates.nc
tie_meteo.nc
tie_geometries.nc
Load OLCI channel information¶
As a first step, you can load one channel with xarray’s function open_dataset. This will help you to understand how the data is structured.
You see that the data of each channel is a two dimensional data array, with rows and columns as dimensions.
olci_xr = xr.open_dataset(olci_dir+'Oa01_radiance.nc')
olci_xr
<xarray.Dataset>
Dimensions: (rows: 4091, columns: 4865)
Dimensions without coordinates: rows, columns
Data variables:
Oa01_radiance (rows, columns) float32 ...
Attributes: (12/16)
absolute_orbit_number: 17105
ac_subsampling_factor: 64
al_subsampling_factor: 1
comment:
contact: eosupport@copernicus.esa.int
creation_time: 2021-08-08T13:01:35Z
... ...
references: S3IPF PDS 004.1 - i2r4 - Product Data Format Spec...
resolution: [ 270 294 ]
source: IPF-OL-1-EO 06.11
start_time: 2021-08-07T08:52:00.193254Z
stop_time: 2021-08-07T08:55:00.160958Z
title: OLCI Level 1b Product, Radiance Oa01 Data Set- rows: 4091
- columns: 4865
- Oa01_radiance(rows, columns)float32...
- ancillary_variables :
- Oa01_radiance_err
- coordinates :
- time_stamp altitude latitude longitude
- long_name :
- TOA radiance for OLCI acquisition band Oa01
- standard_name :
- toa_upwelling_spectral_radiance
- units :
- mW.m-2.sr-1.nm-1
- valid_max :
- 65534
- valid_min :
- 0
[19902715 values with dtype=float32]
- absolute_orbit_number :
- 17105
- ac_subsampling_factor :
- 64
- al_subsampling_factor :
- 1
- comment :
- contact :
- eosupport@copernicus.esa.int
- creation_time :
- 2021-08-08T13:01:35Z
- history :
- 2021-08-08T13:01:35Z: PUGCoreProcessor /data/ipf-s3/workdir42/604551964/JobOrder.604551964.xml
- institution :
- LN1
- netCDF_version :
- 4.2 of Mar 13 2018 10:14:33 $
- product_name :
- S3B_OL_1_EFR____20210807T085200_20210807T085500_20210808T130135_0179_055_278_2340_LN1_O_NT_002.SEN3
- references :
- S3IPF PDS 004.1 - i2r4 - Product Data Format Specification - OLCI Level 1, S3IPF PDS 002 - i1r7 - Product Data Format Specification - Product Structures, S3IPF DPM 002 - i2r5 - Detailed Processing Model - OLCI Level 1
- resolution :
- [ 270 294 ]
- source :
- IPF-OL-1-EO 06.11
- start_time :
- 2021-08-07T08:52:00.193254Z
- stop_time :
- 2021-08-07T08:55:00.160958Z
- title :
- OLCI Level 1b Product, Radiance Oa01 Data Set
Load all channel information into one xarray object¶
The next step is to bring the individual data files together into one xarray data object. You can do this with xarray’s function open_mfdataset. Since all channel files have the same data structure, you can combine all channels by coordinates using the keyword argument by_coords. The result is a xarray object with 21 data variables. Each channel is represented as a data variable.
olci_xr_mf = xr.open_mfdataset(olci_dir+'*_radiance.nc', combine='by_coords')
olci_xr_mf
<xarray.Dataset>
Dimensions: (rows: 4091, columns: 4865)
Dimensions without coordinates: rows, columns
Data variables: (12/21)
Oa01_radiance (rows, columns) float32 dask.array<chunksize=(4091, 4865), meta=np.ndarray>
Oa02_radiance (rows, columns) float32 dask.array<chunksize=(4091, 4865), meta=np.ndarray>
Oa03_radiance (rows, columns) float32 dask.array<chunksize=(4091, 4865), meta=np.ndarray>
Oa04_radiance (rows, columns) float32 dask.array<chunksize=(4091, 4865), meta=np.ndarray>
Oa05_radiance (rows, columns) float32 dask.array<chunksize=(4091, 4865), meta=np.ndarray>
Oa06_radiance (rows, columns) float32 dask.array<chunksize=(4091, 4865), meta=np.ndarray>
... ...
Oa16_radiance (rows, columns) float32 dask.array<chunksize=(4091, 4865), meta=np.ndarray>
Oa17_radiance (rows, columns) float32 dask.array<chunksize=(4091, 4865), meta=np.ndarray>
Oa18_radiance (rows, columns) float32 dask.array<chunksize=(4091, 4865), meta=np.ndarray>
Oa19_radiance (rows, columns) float32 dask.array<chunksize=(4091, 4865), meta=np.ndarray>
Oa20_radiance (rows, columns) float32 dask.array<chunksize=(4091, 4865), meta=np.ndarray>
Oa21_radiance (rows, columns) float32 dask.array<chunksize=(4091, 4865), meta=np.ndarray>
Attributes: (12/16)
absolute_orbit_number: 17105
ac_subsampling_factor: 64
al_subsampling_factor: 1
comment:
contact: eosupport@copernicus.esa.int
creation_time: 2021-08-08T13:01:35Z
... ...
references: S3IPF PDS 004.1 - i2r4 - Product Data Format Spec...
resolution: [ 270 294 ]
source: IPF-OL-1-EO 06.11
start_time: 2021-08-07T08:52:00.193254Z
stop_time: 2021-08-07T08:55:00.160958Z
title: OLCI Level 1b Product, Radiance Oa01 Data Set- rows: 4091
- columns: 4865
- Oa01_radiance(rows, columns)float32dask.array<chunksize=(4091, 4865), meta=np.ndarray>
- ancillary_variables :
- Oa01_radiance_err
- coordinates :
- time_stamp altitude latitude longitude
- long_name :
- TOA radiance for OLCI acquisition band Oa01
- standard_name :
- toa_upwelling_spectral_radiance
- units :
- mW.m-2.sr-1.nm-1
- valid_max :
- 65534
- valid_min :
- 0
Array Chunk Bytes 75.92 MiB 75.92 MiB Shape (4091, 4865) (4091, 4865) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Oa02_radiance(rows, columns)float32dask.array<chunksize=(4091, 4865), meta=np.ndarray>
- ancillary_variables :
- Oa02_radiance_err
- coordinates :
- time_stamp altitude latitude longitude
- long_name :
- TOA radiance for OLCI acquisition band Oa02
- standard_name :
- toa_upwelling_spectral_radiance
- units :
- mW.m-2.sr-1.nm-1
- valid_max :
- 65534
- valid_min :
- 0
Array Chunk Bytes 75.92 MiB 75.92 MiB Shape (4091, 4865) (4091, 4865) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Oa03_radiance(rows, columns)float32dask.array<chunksize=(4091, 4865), meta=np.ndarray>
- ancillary_variables :
- Oa03_radiance_err
- coordinates :
- time_stamp altitude latitude longitude
- long_name :
- TOA radiance for OLCI acquisition band Oa03
- standard_name :
- toa_upwelling_spectral_radiance
- units :
- mW.m-2.sr-1.nm-1
- valid_max :
- 65534
- valid_min :
- 0
Array Chunk Bytes 75.92 MiB 75.92 MiB Shape (4091, 4865) (4091, 4865) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Oa04_radiance(rows, columns)float32dask.array<chunksize=(4091, 4865), meta=np.ndarray>
- ancillary_variables :
- Oa04_radiance_err
- coordinates :
- time_stamp altitude latitude longitude
- long_name :
- TOA radiance for OLCI acquisition band Oa04
- standard_name :
- toa_upwelling_spectral_radiance
- units :
- mW.m-2.sr-1.nm-1
- valid_max :
- 65534
- valid_min :
- 0
Array Chunk Bytes 75.92 MiB 75.92 MiB Shape (4091, 4865) (4091, 4865) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Oa05_radiance(rows, columns)float32dask.array<chunksize=(4091, 4865), meta=np.ndarray>
- ancillary_variables :
- Oa05_radiance_err
- coordinates :
- time_stamp altitude latitude longitude
- long_name :
- TOA radiance for OLCI acquisition band Oa05
- standard_name :
- toa_upwelling_spectral_radiance
- units :
- mW.m-2.sr-1.nm-1
- valid_max :
- 65534
- valid_min :
- 0
Array Chunk Bytes 75.92 MiB 75.92 MiB Shape (4091, 4865) (4091, 4865) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Oa06_radiance(rows, columns)float32dask.array<chunksize=(4091, 4865), meta=np.ndarray>
- ancillary_variables :
- Oa06_radiance_err
- coordinates :
- time_stamp altitude latitude longitude
- long_name :
- TOA radiance for OLCI acquisition band Oa06
- standard_name :
- toa_upwelling_spectral_radiance
- units :
- mW.m-2.sr-1.nm-1
- valid_max :
- 65534
- valid_min :
- 0
Array Chunk Bytes 75.92 MiB 75.92 MiB Shape (4091, 4865) (4091, 4865) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Oa07_radiance(rows, columns)float32dask.array<chunksize=(4091, 4865), meta=np.ndarray>
- ancillary_variables :
- Oa07_radiance_err
- coordinates :
- time_stamp altitude latitude longitude
- long_name :
- TOA radiance for OLCI acquisition band Oa07
- standard_name :
- toa_upwelling_spectral_radiance
- units :
- mW.m-2.sr-1.nm-1
- valid_max :
- 65534
- valid_min :
- 0
Array Chunk Bytes 75.92 MiB 75.92 MiB Shape (4091, 4865) (4091, 4865) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Oa08_radiance(rows, columns)float32dask.array<chunksize=(4091, 4865), meta=np.ndarray>
- ancillary_variables :
- Oa08_radiance_err
- coordinates :
- time_stamp altitude latitude longitude
- long_name :
- TOA radiance for OLCI acquisition band Oa08
- standard_name :
- toa_upwelling_spectral_radiance
- units :
- mW.m-2.sr-1.nm-1
- valid_max :
- 65534
- valid_min :
- 0
Array Chunk Bytes 75.92 MiB 75.92 MiB Shape (4091, 4865) (4091, 4865) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Oa09_radiance(rows, columns)float32dask.array<chunksize=(4091, 4865), meta=np.ndarray>
- ancillary_variables :
- Oa09_radiance_err
- coordinates :
- time_stamp altitude latitude longitude
- long_name :
- TOA radiance for OLCI acquisition band Oa09
- standard_name :
- toa_upwelling_spectral_radiance
- units :
- mW.m-2.sr-1.nm-1
- valid_max :
- 65534
- valid_min :
- 0
Array Chunk Bytes 75.92 MiB 75.92 MiB Shape (4091, 4865) (4091, 4865) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Oa10_radiance(rows, columns)float32dask.array<chunksize=(4091, 4865), meta=np.ndarray>
- ancillary_variables :
- Oa10_radiance_err
- coordinates :
- time_stamp altitude latitude longitude
- long_name :
- TOA radiance for OLCI acquisition band Oa10
- standard_name :
- toa_upwelling_spectral_radiance
- units :
- mW.m-2.sr-1.nm-1
- valid_max :
- 65534
- valid_min :
- 0
Array Chunk Bytes 75.92 MiB 75.92 MiB Shape (4091, 4865) (4091, 4865) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Oa11_radiance(rows, columns)float32dask.array<chunksize=(4091, 4865), meta=np.ndarray>
- ancillary_variables :
- Oa11_radiance_err
- coordinates :
- time_stamp altitude latitude longitude
- long_name :
- TOA radiance for OLCI acquisition band Oa11
- standard_name :
- toa_upwelling_spectral_radiance
- units :
- mW.m-2.sr-1.nm-1
- valid_max :
- 65534
- valid_min :
- 0
Array Chunk Bytes 75.92 MiB 75.92 MiB Shape (4091, 4865) (4091, 4865) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Oa12_radiance(rows, columns)float32dask.array<chunksize=(4091, 4865), meta=np.ndarray>
- ancillary_variables :
- Oa12_radiance_err
- coordinates :
- time_stamp altitude latitude longitude
- long_name :
- TOA radiance for OLCI acquisition band Oa12
- standard_name :
- toa_upwelling_spectral_radiance
- units :
- mW.m-2.sr-1.nm-1
- valid_max :
- 65534
- valid_min :
- 0
Array Chunk Bytes 75.92 MiB 75.92 MiB Shape (4091, 4865) (4091, 4865) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Oa13_radiance(rows, columns)float32dask.array<chunksize=(4091, 4865), meta=np.ndarray>
- ancillary_variables :
- Oa13_radiance_err
- coordinates :
- time_stamp altitude latitude longitude
- long_name :
- TOA radiance for OLCI acquisition band Oa13
- standard_name :
- toa_upwelling_spectral_radiance
- units :
- mW.m-2.sr-1.nm-1
- valid_max :
- 65534
- valid_min :
- 0
Array Chunk Bytes 75.92 MiB 75.92 MiB Shape (4091, 4865) (4091, 4865) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Oa14_radiance(rows, columns)float32dask.array<chunksize=(4091, 4865), meta=np.ndarray>
- ancillary_variables :
- Oa14_radiance_err
- coordinates :
- time_stamp altitude latitude longitude
- long_name :
- TOA radiance for OLCI acquisition band Oa14
- standard_name :
- toa_upwelling_spectral_radiance
- units :
- mW.m-2.sr-1.nm-1
- valid_max :
- 65534
- valid_min :
- 0
Array Chunk Bytes 75.92 MiB 75.92 MiB Shape (4091, 4865) (4091, 4865) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Oa15_radiance(rows, columns)float32dask.array<chunksize=(4091, 4865), meta=np.ndarray>
- ancillary_variables :
- Oa15_radiance_err
- coordinates :
- time_stamp altitude latitude longitude
- long_name :
- TOA radiance for OLCI acquisition band Oa15
- standard_name :
- toa_upwelling_spectral_radiance
- units :
- mW.m-2.sr-1.nm-1
- valid_max :
- 65534
- valid_min :
- 0
Array Chunk Bytes 75.92 MiB 75.92 MiB Shape (4091, 4865) (4091, 4865) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Oa16_radiance(rows, columns)float32dask.array<chunksize=(4091, 4865), meta=np.ndarray>
- ancillary_variables :
- Oa16_radiance_err
- coordinates :
- time_stamp altitude latitude longitude
- long_name :
- TOA radiance for OLCI acquisition band Oa16
- standard_name :
- toa_upwelling_spectral_radiance
- units :
- mW.m-2.sr-1.nm-1
- valid_max :
- 65534
- valid_min :
- 0
Array Chunk Bytes 75.92 MiB 75.92 MiB Shape (4091, 4865) (4091, 4865) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Oa17_radiance(rows, columns)float32dask.array<chunksize=(4091, 4865), meta=np.ndarray>
- ancillary_variables :
- Oa17_radiance_err
- coordinates :
- time_stamp altitude latitude longitude
- long_name :
- TOA radiance for OLCI acquisition band Oa17
- standard_name :
- toa_upwelling_spectral_radiance
- units :
- mW.m-2.sr-1.nm-1
- valid_max :
- 65534
- valid_min :
- 0
Array Chunk Bytes 75.92 MiB 75.92 MiB Shape (4091, 4865) (4091, 4865) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Oa18_radiance(rows, columns)float32dask.array<chunksize=(4091, 4865), meta=np.ndarray>
- ancillary_variables :
- Oa18_radiance_err
- coordinates :
- time_stamp altitude latitude longitude
- long_name :
- TOA radiance for OLCI acquisition band Oa18
- standard_name :
- toa_upwelling_spectral_radiance
- units :
- mW.m-2.sr-1.nm-1
- valid_max :
- 65534
- valid_min :
- 0
Array Chunk Bytes 75.92 MiB 75.92 MiB Shape (4091, 4865) (4091, 4865) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Oa19_radiance(rows, columns)float32dask.array<chunksize=(4091, 4865), meta=np.ndarray>
- ancillary_variables :
- Oa19_radiance_err
- coordinates :
- time_stamp altitude latitude longitude
- long_name :
- TOA radiance for OLCI acquisition band Oa19
- standard_name :
- toa_upwelling_spectral_radiance
- units :
- mW.m-2.sr-1.nm-1
- valid_max :
- 65534
- valid_min :
- 0
Array Chunk Bytes 75.92 MiB 75.92 MiB Shape (4091, 4865) (4091, 4865) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Oa20_radiance(rows, columns)float32dask.array<chunksize=(4091, 4865), meta=np.ndarray>
- ancillary_variables :
- Oa20_radiance_err
- coordinates :
- time_stamp altitude latitude longitude
- long_name :
- TOA radiance for OLCI acquisition band Oa20
- standard_name :
- toa_upwelling_spectral_radiance
- units :
- mW.m-2.sr-1.nm-1
- valid_max :
- 65534
- valid_min :
- 0
Array Chunk Bytes 75.92 MiB 75.92 MiB Shape (4091, 4865) (4091, 4865) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Oa21_radiance(rows, columns)float32dask.array<chunksize=(4091, 4865), meta=np.ndarray>
- ancillary_variables :
- Oa21_radiance_err
- coordinates :
- time_stamp altitude latitude longitude
- long_name :
- TOA radiance for OLCI acquisition band Oa21
- standard_name :
- toa_upwelling_spectral_radiance
- units :
- mW.m-2.sr-1.nm-1
- valid_max :
- 65534
- valid_min :
- 0
Array Chunk Bytes 75.92 MiB 75.92 MiB Shape (4091, 4865) (4091, 4865) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray
- absolute_orbit_number :
- 17105
- ac_subsampling_factor :
- 64
- al_subsampling_factor :
- 1
- comment :
- contact :
- eosupport@copernicus.esa.int
- creation_time :
- 2021-08-08T13:01:35Z
- history :
- 2021-08-08T13:01:35Z: PUGCoreProcessor /data/ipf-s3/workdir42/604551964/JobOrder.604551964.xml
- institution :
- LN1
- netCDF_version :
- 4.2 of Mar 13 2018 10:14:33 $
- product_name :
- S3B_OL_1_EFR____20210807T085200_20210807T085500_20210808T130135_0179_055_278_2340_LN1_O_NT_002.SEN3
- references :
- S3IPF PDS 004.1 - i2r4 - Product Data Format Specification - OLCI Level 1, S3IPF PDS 002 - i1r7 - Product Data Format Specification - Product Structures, S3IPF DPM 002 - i2r5 - Detailed Processing Model - OLCI Level 1
- resolution :
- [ 270 294 ]
- source :
- IPF-OL-1-EO 06.11
- start_time :
- 2021-08-07T08:52:00.193254Z
- stop_time :
- 2021-08-07T08:55:00.160958Z
- title :
- OLCI Level 1b Product, Radiance Oa01 Data Set
Example to plot one channel¶
To get an impression of the image, you can simply plot one channel of the xarray object with xarray’s function .plot.imshow(). Let’s plot channel 8 Oa08_radiance. The example shows that you can easily visualize the image itself, but without geographic information.
fig = plt.figure(figsize=(10,6))
olci_xr_mf['Oa08_radiance'].plot.imshow()
<matplotlib.image.AxesImage at 0x7fec37da17c0>

Load OLCI geographic coordinates¶
If you want to georeference your image, you have to load the geographic coordinates file. You can load it as xarray with open_dataset. The file is called geo_coordinates.nc. You see that the file contains three variables: latitude, longitude and altitude.
Let’s store the latitude and longitude data as lat and lon variables respectively.
olci_geo_coords = xr.open_dataset(olci_dir+'geo_coordinates.nc')
olci_geo_coords
<xarray.Dataset>
Dimensions: (rows: 4091, columns: 4865)
Dimensions without coordinates: rows, columns
Data variables:
altitude (rows, columns) float32 ...
latitude (rows, columns) float64 ...
longitude (rows, columns) float64 ...
Attributes: (12/16)
absolute_orbit_number: 17105
ac_subsampling_factor: 64
al_subsampling_factor: 1
comment:
contact: eosupport@copernicus.esa.int
creation_time: 2021-08-08T13:01:35Z
... ...
references: S3IPF PDS 004.1 - i2r4 - Product Data Format Spec...
resolution: [ 270 294 ]
source: IPF-OL-1-EO 06.11
start_time: 2021-08-07T08:52:00.193254Z
stop_time: 2021-08-07T08:55:00.160958Z
title: OLCI Level 1b Product, Geo Coordinates Data Set- rows: 4091
- columns: 4865
- altitude(rows, columns)float32...
- long_name :
- DEM corrected altitude
- standard_name :
- altitude
- units :
- m
- valid_max :
- 9000
- valid_min :
- -1000
[19902715 values with dtype=float32]
- latitude(rows, columns)float64...
- long_name :
- DEM corrected latitude
- standard_name :
- latitude
- units :
- degrees_north
- valid_max :
- 90000000
- valid_min :
- -90000000
[19902715 values with dtype=float64]
- longitude(rows, columns)float64...
- long_name :
- DEM corrected longitude
- standard_name :
- longitude
- units :
- degrees_east
- valid_max :
- 180000000
- valid_min :
- -180000000
[19902715 values with dtype=float64]
- absolute_orbit_number :
- 17105
- ac_subsampling_factor :
- 64
- al_subsampling_factor :
- 1
- comment :
- contact :
- eosupport@copernicus.esa.int
- creation_time :
- 2021-08-08T13:01:35Z
- history :
- 2021-08-08T13:01:35Z: PUGCoreProcessor /data/ipf-s3/workdir42/604551964/JobOrder.604551964.xml
- institution :
- LN1
- netCDF_version :
- 4.2 of Mar 13 2018 10:14:33 $
- product_name :
- S3B_OL_1_EFR____20210807T085200_20210807T085500_20210808T130135_0179_055_278_2340_LN1_O_NT_002.SEN3
- references :
- S3IPF PDS 004.1 - i2r4 - Product Data Format Specification - OLCI Level 1, S3IPF PDS 002 - i1r7 - Product Data Format Specification - Product Structures, S3IPF DPM 002 - i2r5 - Detailed Processing Model - OLCI Level 1
- resolution :
- [ 270 294 ]
- source :
- IPF-OL-1-EO 06.11
- start_time :
- 2021-08-07T08:52:00.193254Z
- stop_time :
- 2021-08-07T08:55:00.160958Z
- title :
- OLCI Level 1b Product, Geo Coordinates Data Set
lat = olci_geo_coords.latitude.data
lon = olci_geo_coords.longitude.data
Select OLCI channels for a true colour composite¶
Depending on the combination of different OLCI channels, your composite might highlight specific phenomena. The channel combination for a True Colour image could be:
Red:
Oa08_radianceGreen:
Oa06_radianceBlue:
Oa04_radiance
Let’s define a function called select_channels_for_rgb, which makes the channel selection more flexible. The function returns the three bands individually.
def select_channels_for_rgb(xarray, red_channel, green_channel, blue_channel):
"""
Selects the channels / bands of a multi-dimensional xarray for red, green and blue composites.
Parameters:
xarray(xarray Dataset): xarray Dataset object that stores the different channels / bands.
red_channel(str): Name of red channel to be selected
green_channel(str): Name of green channel to be selected
blue_channel(str): Name of blue channel to be selected
Returns:
Three xarray DataArray objects with selected channels / bands
"""
return xarray[red_channel], xarray[green_channel], xarray[blue_channel]
red, green, blue = select_channels_for_rgb(olci_xr_mf, 'Oa08_radiance', 'Oa06_radiance', 'Oa04_radiance')
red
<xarray.DataArray 'Oa08_radiance' (rows: 4091, columns: 4865)>
dask.array<open_dataset-b8fb34b5e082f5b0069d93edb3a6dedeOa08_radiance, shape=(4091, 4865), dtype=float32, chunksize=(4091, 4865), chunktype=numpy.ndarray>
Dimensions without coordinates: rows, columns
Attributes:
ancillary_variables: Oa08_radiance_err
coordinates: time_stamp altitude latitude longitude
long_name: TOA radiance for OLCI acquisition band Oa08
standard_name: toa_upwelling_spectral_radiance
units: mW.m-2.sr-1.nm-1
valid_max: 65534
valid_min: 0- rows: 4091
- columns: 4865
- dask.array<chunksize=(4091, 4865), meta=np.ndarray>
Array Chunk Bytes 75.92 MiB 75.92 MiB Shape (4091, 4865) (4091, 4865) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - ancillary_variables :
- Oa08_radiance_err
- coordinates :
- time_stamp altitude latitude longitude
- long_name :
- TOA radiance for OLCI acquisition band Oa08
- standard_name :
- toa_upwelling_spectral_radiance
- units :
- mW.m-2.sr-1.nm-1
- valid_max :
- 65534
- valid_min :
- 0
Advanced image processing - Normalization and histogram equalization¶
Normalization¶
A common operation in image processing is the normalization of data values. Normalization changes the range of pixel intesity and can improve the constrast. Let’s define a function called normalize , which normalizes a numpy array into a scale between 0.0 and 1.0.
def normalize(array):
"""
Normalizes a numpy array / xarray DataArray object value to values between 0 and 1.
Parameters:
xarray(numpy array or xarray DataArray): xarray DataArray or numpy array object.
Returns:
Normalized array
"""
array_min, array_max = array.min(), array.max()
return ((array - array_min)/(array_max - array_min))
You can now apply the function normalizeto each RGB channel. At the end, you can bring the three channels together into one rgb array with numpy.dstack. By verifying the shape of the resulting array, you see that the rgb array has now three dimensions.
redn = normalize(red)
greenn = normalize(green)
bluen = normalize(blue)
rgb = np.dstack((redn, greenn, bluen))
rgb.shape
(4091, 4865, 3)
You can now plot the RGB image to see if maybe further image processing methods could be applied. If you visualize the image with plt.imshow(), you see that the contrast is not very strong. A further step is to sharpen the contrast with the help of a Histogram equalization.
fig = plt.figure(figsize=(10,6))
plt.imshow(rgb)
<matplotlib.image.AxesImage at 0x7feb1160e1c0>

Histogram equalization¶
Histogram equalization is a method in image processing that adjusts the contrast using the image’s histogram. Python’s skikit-learn library has useful tools to make a histogram equalization quite straighforward. The skimage library provides a function called exposure.equalize_adapthist() which can be applied to the RGB data array.
rgb = exposure.equalize_adapthist(rgb)
You can again plot the RGB image with plt.imshow(). You can see that the histogram equalization improved the contrast of the image.
fig = plt.figure(figsize=(10,6))
plt.imshow(rgb)
<matplotlib.image.AxesImage at 0x7feb11575940>
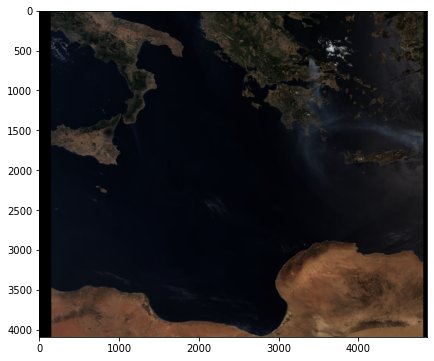
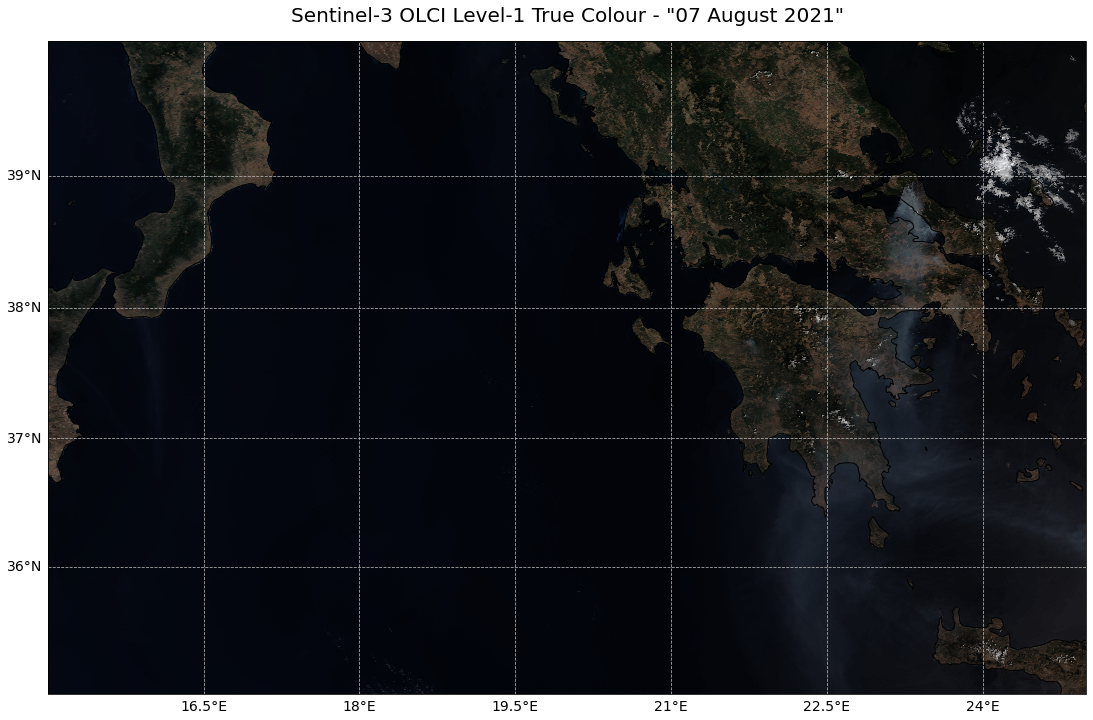
Visualise a georeferenced true colour composite¶
The final step is to georeference the true colour composite image. Therefore, you need the extracted lat and lon information extracted above.
Let’s plot the image with matplotlib’s pcolormesh function. To be able to visualize the array as RGB, you have to first map the RGB array to a colour array.
mesh_rgb = rgb[:, :, :]
colorTuple = mesh_rgb.reshape((mesh_rgb.shape[0] * mesh_rgb.shape[1]), 3)
colorTuple = np.insert(colorTuple, 3, 1.0, axis=1)
The last step is to visualize the color array and add additional information, e.g. gridlines and coastlines. As you want to reuse the plotting code again, you can define a function called visualize_s3_pcolormesh.
def visualize_s3_pcolormesh(color_array, array, latitude, longitude, title):
"""
Visualizes a numpy array (Sentinel-3 data) with matplotlib's 'pcolormesh' function as composite.
Parameters:
color_array (numpy MaskedArray): any numpy MaskedArray, e.g. loaded with the NetCDF library and the Dataset function
longitude (numpy Array): array with longitude values
latitude (numpy Array) : array with latitude values
title (str): title of the resulting plot
"""
fig=plt.figure(figsize=(20, 12))
ax=plt.axes(projection=ccrs.Mercator())
ax.coastlines()
ax.set_extent([15, 25, 35, 40]) # Mediterranean
gl = ax.gridlines(draw_labels=True, linestyle='--')
gl.xlabels_top=False
gl.ylabels_right=False
gl.xformatter=LONGITUDE_FORMATTER
gl.yformatter=LATITUDE_FORMATTER
gl.xlabel_style={'size':14}
gl.ylabel_style={'size':14}
img1 = plt.pcolormesh(longitude, latitude, array*np.nan, color=colorTuple,
clip_on = True,
edgecolors=None,
zorder=0,
transform=ccrs.PlateCarree()
)
ax.set_title(title, fontsize=20, pad=20.0)
plt.show()
visualize_s3_pcolormesh(color_array=colorTuple,
array=red,
latitude=lat,
longitude=lon,
title='Sentinel-3 OLCI Level-1 True Colour - "07 August 2021"')
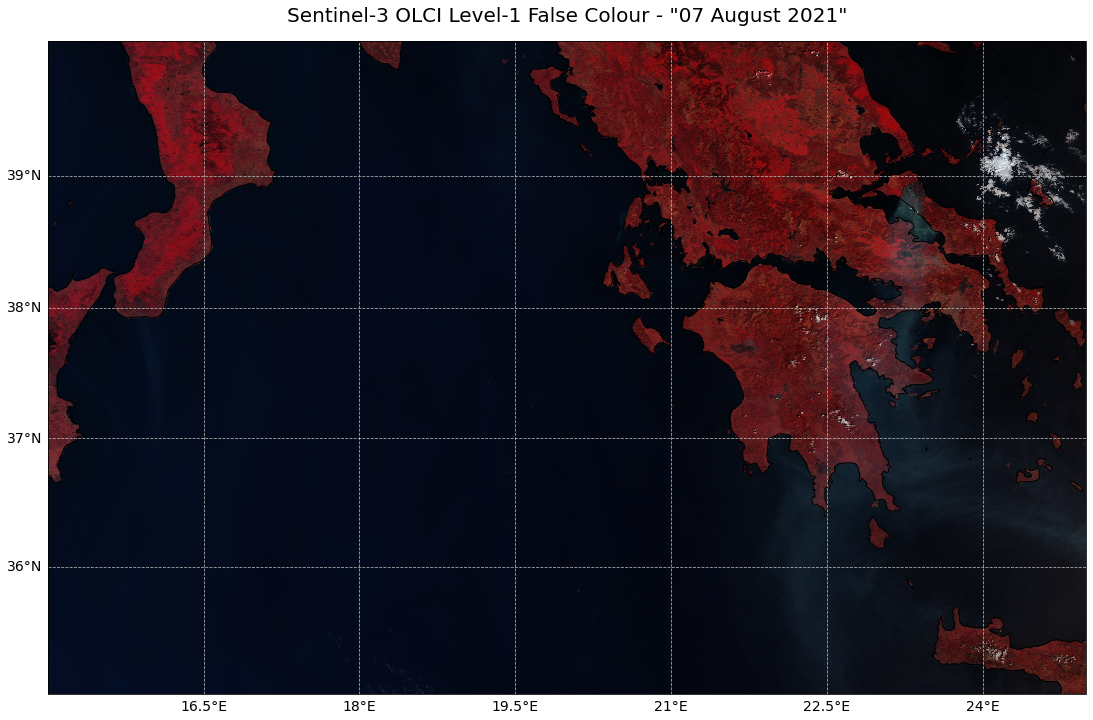
Create a false colour composite and plot it as georeferenced image¶
A false colour image can be applied if specific phenomena should be highlighted. A channel combination for Sentinel-3 OLCI data could be:
Red:
Oa17_radianceGreen:
Oa05_radianceBlue:
Oa02_radiance
This selection is advantageous to identify burnt areas and wildfires. The combination highlights healthy vegetation as red and burnt areas as black.
Let’s repeat the steps from above to visualize the image as False Colour composite.
Select RGB channels¶
The first step is to select the channels Oa17_radiance, Oa05_radiance and Oa02_radiance as red, green and blue channels respectively.
red_fc, green_fc, blue_fc = select_channels_for_rgb(olci_xr_mf, 'Oa17_radiance', 'Oa05_radiance', 'Oa02_radiance')
Normalize¶
You also want to normalize these channels and stack them afterwards into a three-dimensional array.
redn_fc = normalize(red_fc)
greenn_fc = normalize(green_fc)
bluen_fc = normalize(blue_fc)
rgb_fc = np.dstack((redn_fc, greenn_fc, bluen_fc))
Histogram equalization¶
Apply histogram equalization.
rgb_fc = exposure.equalize_adapthist(rgb_fc)
Let’s have a sneak peek at the un-georeferenced image with plt.imshow()
fig = plt.figure(figsize=(10,6))
plt.imshow(rgb_fc)
<matplotlib.image.AxesImage at 0x7feb1117fe20>

Visualize with function visualize_s3_pcolormesh¶
mesh_rgb = rgb_fc[:, :, :]
colorTuple = mesh_rgb.reshape((mesh_rgb.shape[0] * mesh_rgb.shape[1]), 3)
colorTuple = np.insert(colorTuple, 3, 1.0, axis=1)
visualize_s3_pcolormesh(color_array=colorTuple,
array=red,
latitude=lat,
longitude=lon,
title='Sentinel-3 OLCI Level-1 False Colour - "07 August 2021"',
)
References¶
Copernicus Sentinel data 2021
Some code in this notebook was adapted from the following source
copyright: 2022, EUMETSAT
license: MIT
retrieved: 2022-06-28 by Sabrina Szeto
Return to the case study
Monitoring active fires with next-generation satellites: Mediterranean Fires Case Study
False colour composites
